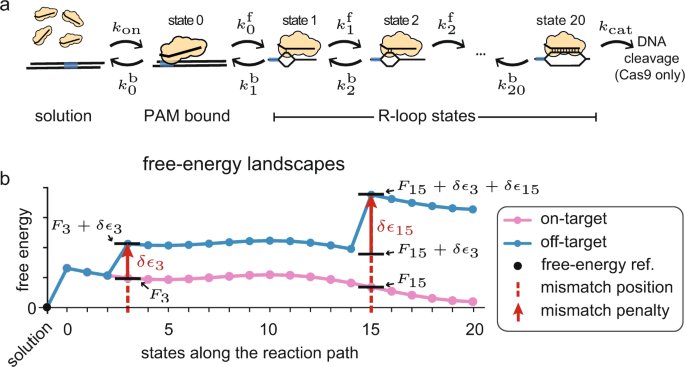
A kinetic model predicts SpCas9 activity, improves off-target classification, and reveals the physical basis of targeting fidelity

PDF) A kinetic model predicts SpCas9 activity, improves off-target classification, and reveals the physical basis of targeting fidelity

A deep learning-based model DeepSpCas9 to predict SpCas9 activity

Massively parallel kinetic profiling of natural and engineered CRISPR nucleases
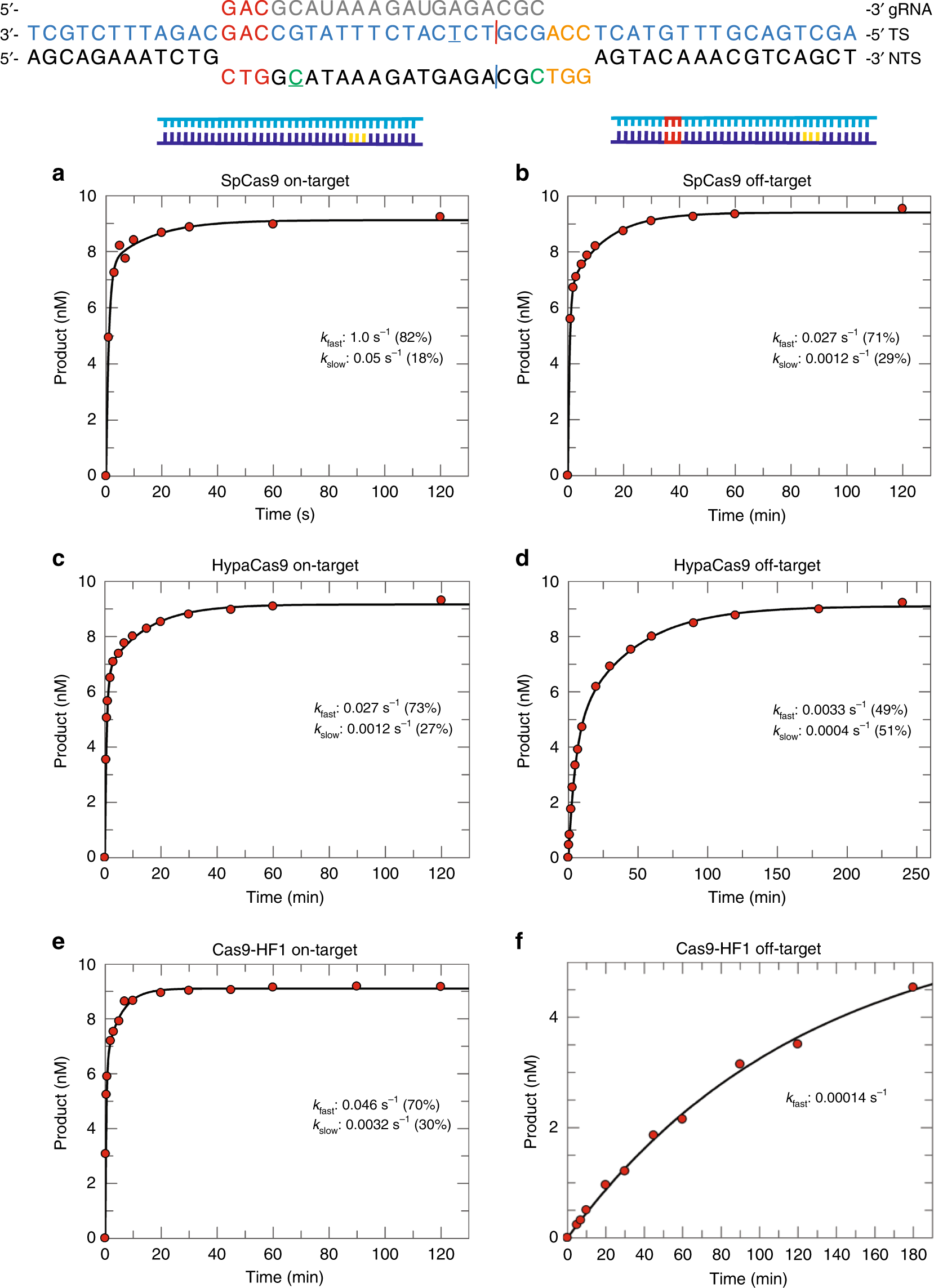
Engineered CRISPR/Cas9 enzymes improve discrimination by slowing DNA cleavage to allow release of off-target DNA

Altered DNA repair pathway engagement by engineered CRISPR-Cas9 nucleases

A kinetic model improves off-target predictions and reveals the physical basis of SpCas9 fidelity

A quantitative model for the dynamics of target recognition and off-target rejection by the CRISPR-Cas Cascade complex
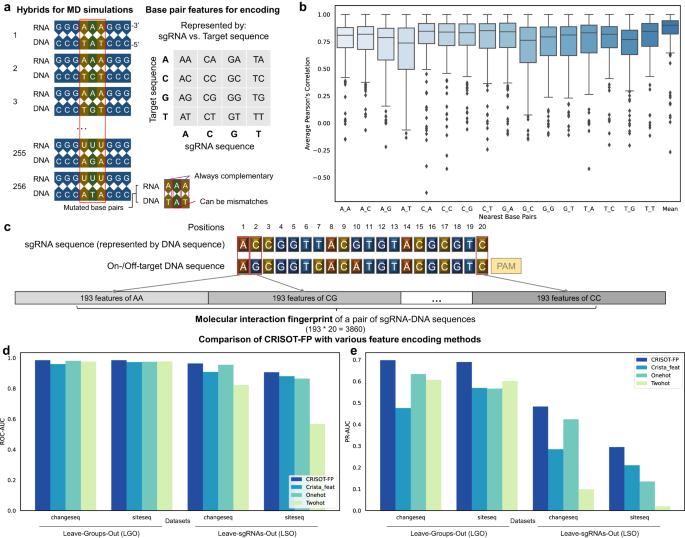
Genome-wide CRISPR off-target prediction and optimization using RNA-DNA interaction fingerprints

Negative DNA supercoiling induces genome-wide Cas9 off-target activity - ScienceDirect

Prediction of off-target effect and gRNA specificity with MOFF a A

PDF) Genome-wide CRISPR off-target prediction and optimization using RNA-DNA interaction fingerprints

A kinetic model predicts SpCas9 activity, improves off-target classification, and reveals the physical basis of targeting fidelity
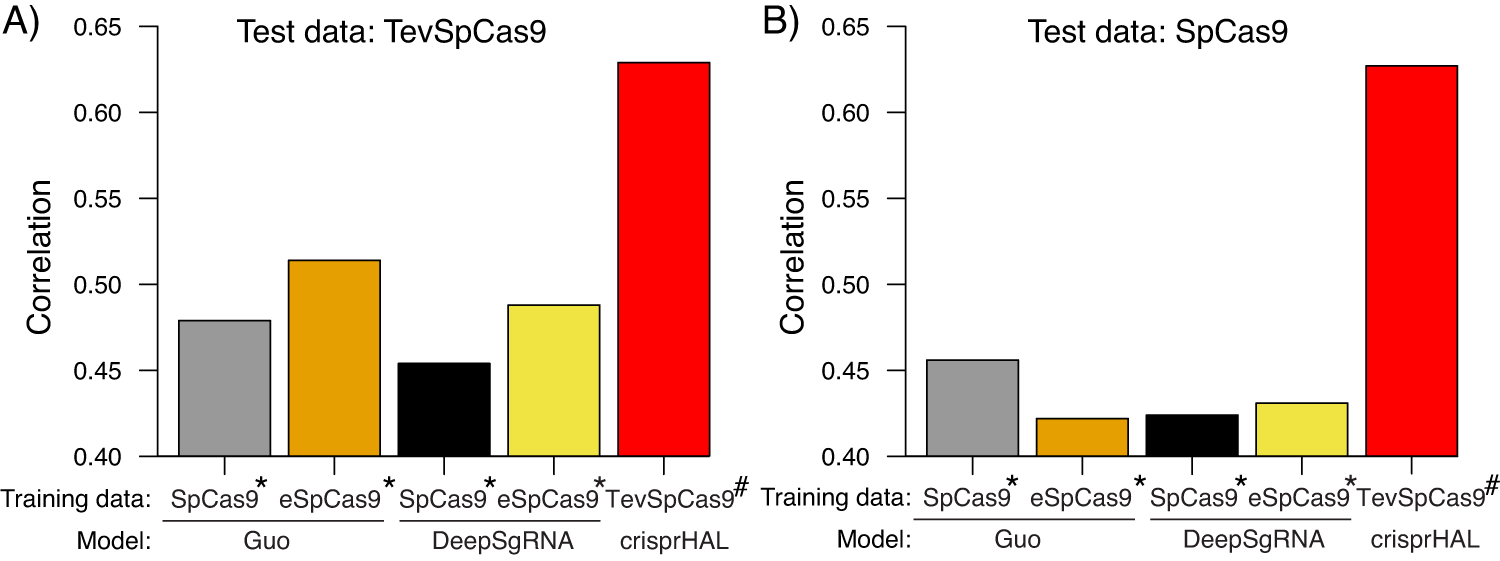
A generalizable Cas9/sgRNA prediction model using machine transfer learning with small high-quality datasets

Massively parallel evaluation and computational prediction of the activities and specificities of 17 small Cas9s

Prediction of the sequence-specific cleavage activity of Cas9 variants









